| Organism Information |
| Organism Name | Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) |
| Clinical Implication | Pathogenic |
| Domain | Bacteria |
| Classification | Phylum : Proteobacteria
Class : EpsilonProteobacteria
Orders : Campylobacterales
Family : Campylobacteraceae
Genus : Campylobacter
Species : jejuni
Subspecies : jejuni
Strain : ATCC 700819 / NCTC 11168 |
| Taxonomic ID (NCBI) | 192222 |
| Genome Information |
| Gene Bank | AL111168 |
| EMBL | AL111168 |
| Gene Information |
| Gene Name | pglB |
| NCBI Gene ID | 905417 |
| Protein information |
| Protein Name | PglB |
| UniProtKB/ SwissProt ID | Q0P9C8 |
| NCBI Ref Seq | YP_002344519.1 |
| UniProtKB Sequence | >sp|Q0P9C8|PGLB_CAMJE Undecaprenyl-diphosphooligosaccharide--protein glycotransferase OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) GN=pglB PE=1 SV=1
MLKKEYLKNPYLVLFAMIVLAYVFSVFCRFYWVWWASEFNEYFFNNQLMIISNDGYAFAE
GARDMIAGFHQPNDLSYYGSSLSTLTYWLYKITPFSFESIILYMSTFLSSLVVIPIILLA
NEYKRPLMGFVAALLASVANSYYNRTMSGYYDTDMLVIVLPMFILFFMVRMILKKDFFSL
IALPLFIGIYLWWYPSSYTLNVALIGLFLIYTLIFHRKEKIFYIAVILSSLTLSNIAWFY
QSAIIVILFALFALEQKRLNFMIIGILGSATLIFLILSGGVDPILYQLKFYIFRSDESAN
LTQGFMYFNVNQTIQEVENVDFSEFMRRISGSEIVFLFSLFGFVWLLRKHKSMIMALPIL
VLGFLALKGGLRFTIYSVPVMALGFGFLLSEFKAILVKKYSQLTSNVCIVFATILTLAPV
FIHIYNYKAPTVFSQNEASLLNQLKNIANREDYVVTWWDYGYPVRYYSDVKTLVDGGKHL
GKDNFFPSFSLSKDEQAAANMARLSVEYTEKSFYAPQNDILKSDILQAMMKDYNQSNVDL
FLASLSKPDFKIDTPKTRDIYLYMPARMSLIFSTVASFSFINLDTGVLDKPFTFSTAYPL
DVKNGEIYLSNGVVLSDDFRSFKIGDNVVSVNSIVEINSIKQGEYKITPIDDKAQFYIFY
LKDSAIPYAQFILMDKTMFNSAYVQMFFLGNYDKNLFDLVINSRDAKVFKLKI
|
| EMBL CDS | CAL35243.1 |
| Sequence length | 713 AA |
| Subcellular Location | Membrane (Integral component of membrane) |
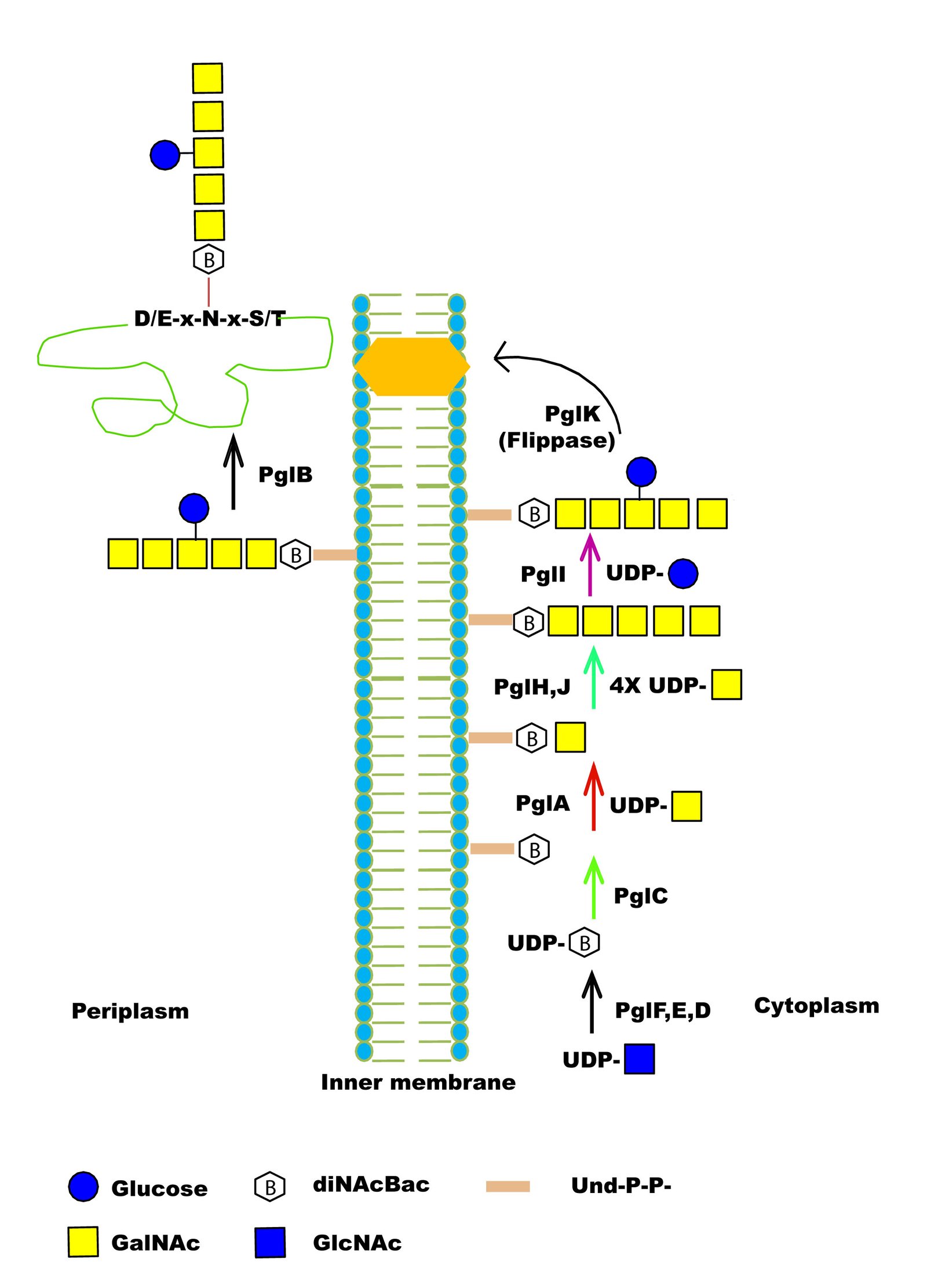
| Function in Native Organism | 1) The PglB is OSTase which transfer an heptasaccharide to N of D/E-N-X-T/S consensus sequence of the protein in Campylobacter jejuni. |
| String | 192222.Cj1126c. |
| Potential Application | 1) PglB can be used to create artificial glycopeptides because it has relaxed substrate specificity of accepting peptide substrates over the full-length protein.
2) PglB can transfer undecaprenyl pyrophosphate-linked saccharides of various lengths (2?7 saccharides) adds to the promise of using PglB in the synthesis of diverse glycopeptide products. |
| Additional Information | 1) PglB requires an acetamido group at the C-2.
2) R331 of C.lari and R328 of C.jejuni form a salt bridge with acidic amino acid.
3) PglB can transfer several structurally different O-antigen saccharides to protein.
|
| Glycosyltransferase Information |
| Glycosylation Type | N- (Asn) linked |
| CAZY Family | GT66 |
| EC Number (BRENDA) | 2.4.1.- |
| Mechanism of Glycan Transfer | En bloc |
| Acceptor specificity Sequon_1 | Asp/Glu- Asn-Xaa-Ser/Thr |
| Donor Type | UndPP-GalNAc- 1,4-GalNAc- 1,4-(Glc 1,3)-Gal-NAc- 1,4- GalNAc- 1,4-GalNAc- 1,3-Bac |
| Donor Specificity | Lipid linked sugars |
| Accessory GT ID | ProGT10.1, ProGT10.2, ProGT10.3, ProGT10.4, ProGT10.5, ProGT10.6, ProGT10.7, ProGT10.8, ProGT10.9 |
| Glycan Information |
| Glycan transferred | Heptasaccharide (GalNAc- 1,4-GalNAc- 1,4-(Glc 1,3)-Gal-NAc- 1,4- GalNAc- 1,4-GalNAc- 1,3-Bac, where Bac is bacillosamine(2,4-diacetamido-2,4,6-trideoxygluose)) |
| Method of Glycan Indentification | NMR and GC MS |
| Experimental_strategies | In vivo and In vitro |
| Acceptor Subtrate Information |
| Acceptor Substrate name | CgpA |
| ProGPdb ID | ProGP218 |
| Acceptor Substrate name | Cj0114 |
| ProGPdb ID | ProGP219 |
| Acceptor Substrate name | Cj0200c |
| ProGPdb ID | ProGP220 |
| Acceptor Substrate name | Cj1496c |
| ProGPdb ID | ProGP221 |
| Acceptor Substrate name | PEB3 |
| ProGPdb ID | ProGP222 |
| Acceptor Substrate name | ZnuA |
| ProGPdb ID | ProGP223 |
| Acceptor Substrate name | AcrA |
| ProGPdb ID | ProGP218 |
| Litrature |
| Year Of Validation | 2002 |
| Reference | Wacker, M., Linton, D., Hitchen, P.G., Nita-Lazar, M., Haslam, S.M., North, S.J., Panico, M., Morris, H.R., Dell, A., Wren, B.W. and Aebi, M., 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into Escherichia coli. Science, 298(5599), pp.1790-1793.
|
| Corresponding Author | Institute for Biological Sciences, National Research Council of Canada, 100 Sussex Dr., Ottawa, Ontario K1A 0R6, Canada.
|
| Reference | Glover, K.J., Weerapana, E., Numao, S. and Imperiali, B., 2005. Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chemistry & biology, 12(12), pp.1311-1316.
|
| Corresponding Author | Department of Chemistry and Biology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139, USA.
|
| Reference | Kowarik, M., Young, N.M., Numao, S., Schulz, B.L., Hug, I., Callewaert, N., Mills, D.C., Watson, D.C., Hernandez, M., Kelly, J.F. and Wacker, M., 2006. Definition of the bacterial N?glycosylation site consensus sequence. The EMBO journal, 25(9), pp.1957-1966.
|
| Corresponding Author | Institute of Microbiology, Department of Biology, Swiss Federal Institute of Technology Zurich, ETH Hönggerberg, Switzerland
|
| Reference | Ihssen, J., Kowarik, M., Wiesli, L., Reiss, R., Wacker, M. and Thöny-Meyer, L., 2012. Structural insights from random mutagenesis of Campylobacter jejunioligosaccharyltransferase PglB. BMC biotechnology, 12(1), pp.1-13.
|
| Corresponding Author | Empa, Swiss Federal Laboratories for Materials Science and Technology, Laboratory for Biomaterials, CH-9014, St, Gallen, Switzerland.
|
| Reference | Jaffee, M.B. and Imperiali, B., 2013. Optimized protocol for expression and purification of membrane-bound PglB, a bacterial oligosaccharyl transferase. Protein expression and purification, 89(2), pp.241-250.
|
| Corresponding Author | Department of Biology and Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge
|
| Reference | Ollis, A.A., Zhang, S., Fisher, A.C. and DeLisa, M.P., 2014. Engineered oligosaccharyltransferases with greatly relaxed acceptor-site specificity. Nature chemical biology, 10(10), pp.816-822.
|
| Corresponding Author | School of Chemical and Biomolecular Engineering, Cornell University, Ithaca, New York, USA
|