| Organism Information |
| Organism Name | Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) (Halobacterium volcanii) |
| Clinical Implication | Non-pathogenic |
| Domain | Archaebacteria |
| Classification | Phylum : Euryarchaeota
Class : Halobaccteria
Orders : Haloferacales
Family : Haloferacaceae
Genus : Haloferax
Species : volcanii
Strain : ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2 |
| Taxonomic ID (NCBI) | 309800 |
| Genome Information |
| Gene Bank | CP001956.1 |
| EMBL | AM922226 |
| Gene Information |
| Gene Name | aglB |
| NCBI Gene ID | 8923906 |
| Protein information |
| Protein Name | AglB |
| UniProtKB/ SwissProt ID | D4GYH4 |
| NCBI Ref Seq | YP_003535577.1 |
| UniProtKB Sequence | >sp|D4GYH4|AGLB_HALVD Dolichyl-monophosphooligosaccharide--protein glycotransferase AglB OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) GN=aglB PE=1 SV=1
MSDEQTKYSPSIAELARDWYHIPVLSTIILVMLWIRLRSYDAFIREGTVFFSGNDAWYHL
RQVEYTVRNWPATMPFDPWTEFPFGRTAGQFGTIYDQLVATAALVVGLGSPSSDLVAKSL
LVAPAVFGALTVIPTYLIGKRLGGRLGGLFGAVILMLLPGTFLQRGLVGFADHNIVEPFF
MGFAVLAIMIALTVADREKPVWELVAARDLDALREPLKWSVLAGVATAIYMWSWPPGILL
VGIFGLFLVLKMASDYVRGRSPEHTAFVGAISMTVTGLLMFIPIEEPGFGVTDFGFLQPL
FSLGVALGAVFLAALARWWESNDVDERYYPAVVGGTMLVGIVLFSLVLPSVFDSIARNFL
RTVGFSAGAATRTISEAQPFLAANVLQSNGQTAVGRIMSEYGFTFFTGALAAVWLVAKPL
VKGGNSRKIGYAVGSLALIGVLFLIPALPAGIGSALGVEPSLVSLTIVTALIVGAVMQAD
YESERLFVLVWAAIITSAAFTQVRFNYYLAVVVAVMNAYLLREALGIDFVGLANVERFDD
ISYGQVAAVVIAVLLILTPVLIIPIQLGNGGVSQTAMQASQTGPGTVTQWDGSLTWMQNN
TPAEGEFGGESNRMEYYGTYEYTDDFDYPDGAYGVMSWWDYGHWITVLGERIPNANPFQG
GATEAANYLLAEDEQQAESVLTSMGDDGEGDQTRYVMVDWQMASTDAKFSAPTVFYDESN
ISRSDFYNPMFRLQEQGEQTTVAAASSLKDQRYYESLMVRLYAYHGSAREASPIVVDWEE
RTSADGSTTFRVTPSDGQAVRTFDNMSAAEEYVANDPTSQIGGIGTFPEERVSALEHYRL
VKSSNSSALRSGSYQRSLISEGNTYGLQPQALVPNNPAWVKTFERVPGATVDGSGAPANT
TVTARVQMRDLTTGTNFTYTQQAQTDADGEFTMTLPYSTTGYDEYGPDNGYTNVSVRAAG
GYAFTGPTSVTGNSTIVSYQAENVAVDEGLVNGAEDGTVQVTLERNEQELDLPGDSSSED
SSSEDGTSDGSQTNESASTSTSASVDASAVSAAA |
| EMBL CDS | CAP58184.1 |
| Sequence length | 1054 AA |
| Subcellular Location | Membrane (Integral component of membrane) |
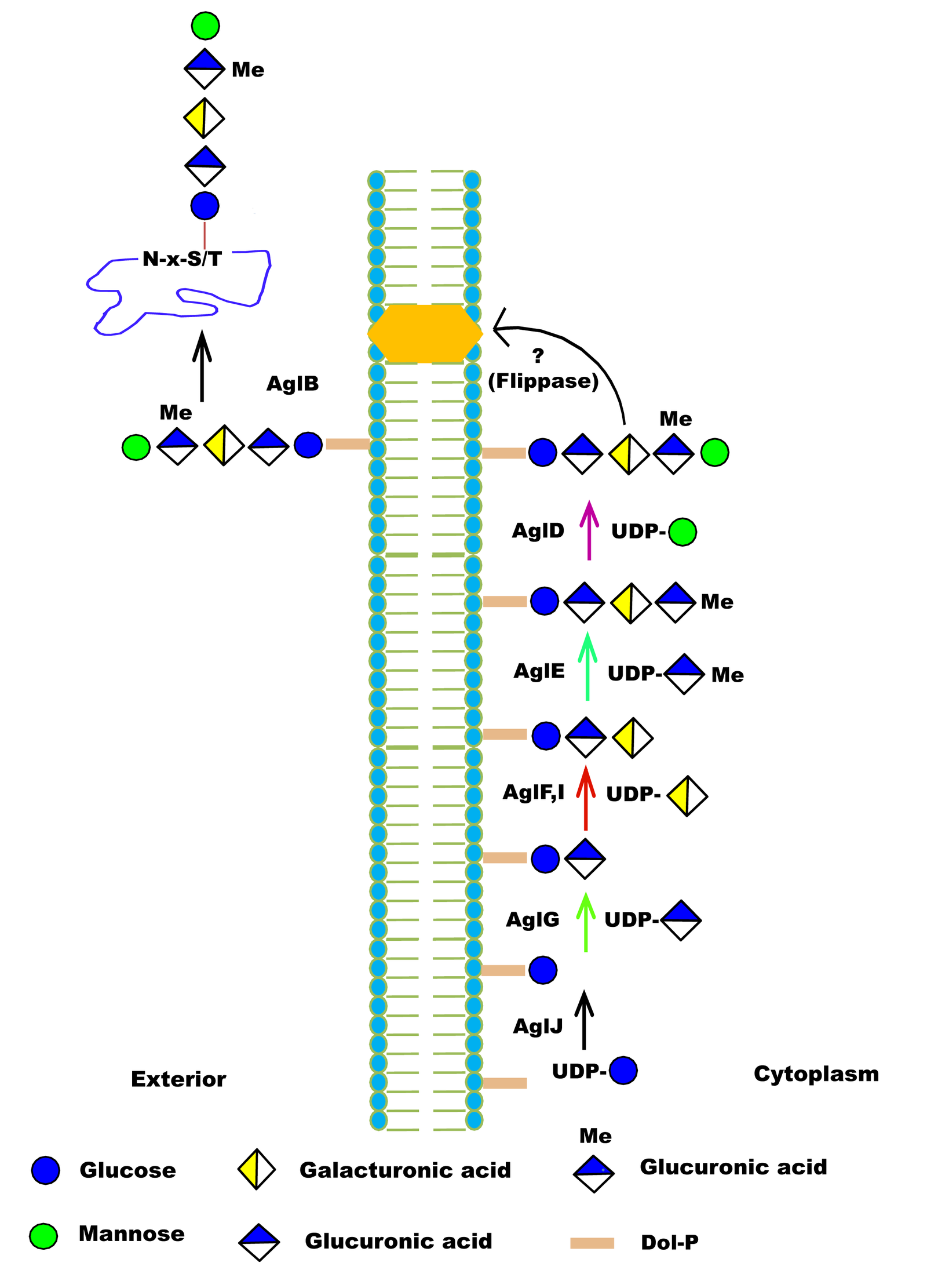
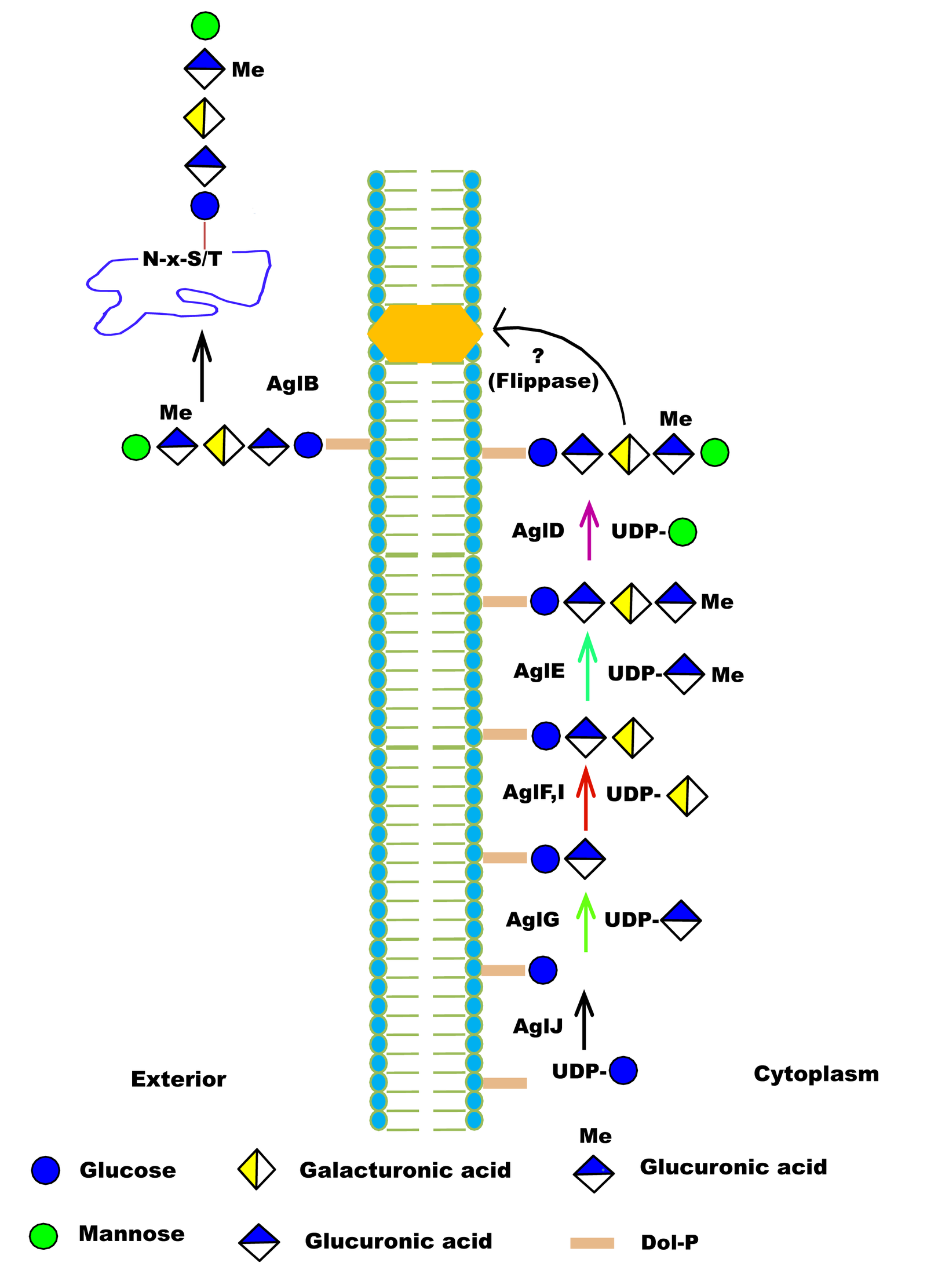
| Function in Native Organism | 1) AglB is OSTase transfer the pentasaccharide to the S-layer protein.
2) For motility of flagella, the presence of the entire pentasaccharide is essential, and for the flagellum assembly requires AglB-dependent glycosylation of FlgA1. |
| String | 309800.HVO_1530. |
| Additional Information | 1) AglB and AglD are involved in assembly of S-layer glycoproteins.
2) H. volcanii AglB shows a relaxed substrate specificity and can transfer truncated glycan to the S-layer glycoprotein, as in aglD deleted cells, similar OSTs also reported in C. jejuni PglB and AglB from the methanoarchaea M. voltae |
| Glycosyltransferase Information |
| Glycosylation Type | N- (Asn) linked |
| CAZY Family | GT66 |
| EC Number (BRENDA) | 2.4.1.- |
| Mechanism of Glycan Transfer | En bloc |
| Acceptor specificity Sequon_1 | Asn-Xaa-Ser |
| Donor Type | DolP-Pentasaccharide |
| Donor Specificity | Lipid linked sugars |
| Accessory GT ID | ProGT15.1, ProGT15.2, ProGT15.3, ProGT15.4, ProGT15.5, ProGT15.6, ProGT15.7, ProGT15.8, ProGT15.9, ProGT15.10, ProGT15.11, ProGT15.12,ProGT15.13, ProGT15.14, ProGT15.15, ProGT15.16, ProGT15.17, ProGT15.18, ProGT15.19, ProGT15.20, ProGT15.21, ProGT15.22 |
| Glycan Information |
| Glycan transferred | Pentasaccharide (2 Monosaccharides (hexose)s, 2 hexuronic acids and a methyl ester of hexuronic acid) |
| Method of Glycan Indentification | In-source collision-induced dissociation (IS-CID), LC-ESI-MS and LC-MS/MS |
| Experimental_strategies | In vivo |
| Acceptor Subtrate Information |
| Acceptor Substrate name | S-layer glycoprotein |
| ProGPdb ID | ProGP71 |
| Acceptor Substrate name | PilA1 |
| ProGPdb ID | ProGP545 |
| Acceptor Substrate name | PilA2 |
| ProGPdb ID | ProGP546 |
| Acceptor Substrate name | PilA3 |
| ProGPdb ID | ProGP547 |
| Acceptor Substrate name | PilA4 |
| ProGPdb ID | ProGP548 |
| Acceptor Substrate name | PilA6 |
| ProGPdb ID | ProGP549 |
| Acceptor Substrate name | FlgA1 |
| ProGPdb ID | ProGP543 |
| Acceptor Substrate name | FlgA2 |
| ProGPdb ID | ProGP544 |
| Litrature |
| Year Of Validation | 2006 |
| Reference | Abu?Qarn, M. and Eichler, J., 2006. Protein N?glycosylation in Archaea: defining Haloferax volcanii genes involved in S?layer glycoprotein glycosylation. Molecular microbiology, 61(2), pp.511-525.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Yurist?Doutsch, S., Abu?Qarn, M., Battaglia, F., Morris, H.R., Hitchen, P.G., Dell, A. and Eichler, J., 2008. aglF, aglG and aglI, novel members of a gene island involved in the N?glycosylation of the Haloferax volcanii S?layer glycoprotein. Molecular microbiology, 69(5), pp.1234-1245.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Kaminski, L., Abu-Qarn, M., Guan, Z., Naparstek, S., Ventura, V.V., Raetz, C.R., Hitchen, P.G., Dell, A. and Eichler, J., 2010. AglJ adds the first sugar of the N-linked pentasaccharide decorating the Haloferax volcanii S-layer glycoprotein. Journal of bacteriology, 192(21), pp.5572-5579.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Yurist?Doutsch, S., Magidovich, H., Ventura, V.V., Hitchen, P.G., Dell, A. and Eichler, J., 2010. N?glycosylation in Archaea: on the coordinated actions of Haloferax volcanii AglF and AglM. Molecular microbiology, 75(4), pp.1047-1058.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Cohen-Rosenzweig, C., Yurist-Doutsch, S. and Eichler, J., 2012. AglS, a novel component of the Haloferax volcanii N-glycosylation pathway, is a dolichol phosphate-mannose mannosyltransferase. Journal of bacteriology, 194(24), pp.6909-6916.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Kaminski, L., Guan, Z., Abu-Qarn, M., Konrad, Z. and Eichler, J., 2012. AglR is required for addition of the final mannose residue of the N-linked glycan decorating the Haloferax volcanii S-layer glycoprotein. Biochimica et Biophysica Acta (BBA)-General Subjects, 1820(10), pp.1664-1670.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Tripepi, M., You, J., Temel, S., Önder, Ö., Brisson, D. and Pohlschröder, M., 2012. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. Journal of bacteriology, 194(18), pp.4876-4887.
|
| Corresponding Author | Department of Biology, University of Pennsylvania, Philadelphia, Pennsylvania, USA
|
| Reference | Arbiv, A., Yurist-Doutsch, S., Guan, Z. and Eichler, J., 2013. AglQ is a novel component of the Haloferax volcanii N-glycosylation pathway. PLoS One, 8(11), p.e81782.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Abu-Qarn, M., Yurist-Doutsch, S., Giordano, A., Trauner, A., Morris, H.R., Hitchen, P., Medalia, O., Dell, A. and Eichler, J., 2007. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. Journal of molecular biology, 374(5), pp.1224-1236.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Kaminski, L., Guan, Z., Yurist-Doutsch, S. and Eichler, J., 2013. Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. MBio, 4(6), pp.e00716-13.
|
| Corresponding Author | Department of Life Sciences, Ben Gurion University, Beersheva 84105, Israel.
|
| Reference | Esquivel, R.N., Schulze, S., Xu, R., Hippler, M. and Pohlschroder, M., 2016. Identification of Haloferax volcanii pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. Journal of Biological Chemistry, 291(20), pp.10602-10614.
|
| Corresponding Author | Department of Biology, University of Pennsylvania, Philadelphia, Pennsylvania, USA
|